sRNAbench Results for job ID: 3ZJI54CCAE8CHON
The results will be stored for 15 days (This will be removed on: Tue 14 May 2019).
If you use the sRNAbench webserver plese check How to Cite.
Download all results as zip file here
---- Mapping parameters and Annotations ----
Number of allowed mismatches: 2 (mm=2)
Alignment type: seed alignment (alignType=n)
Length of the seed: 20 (seed=20)
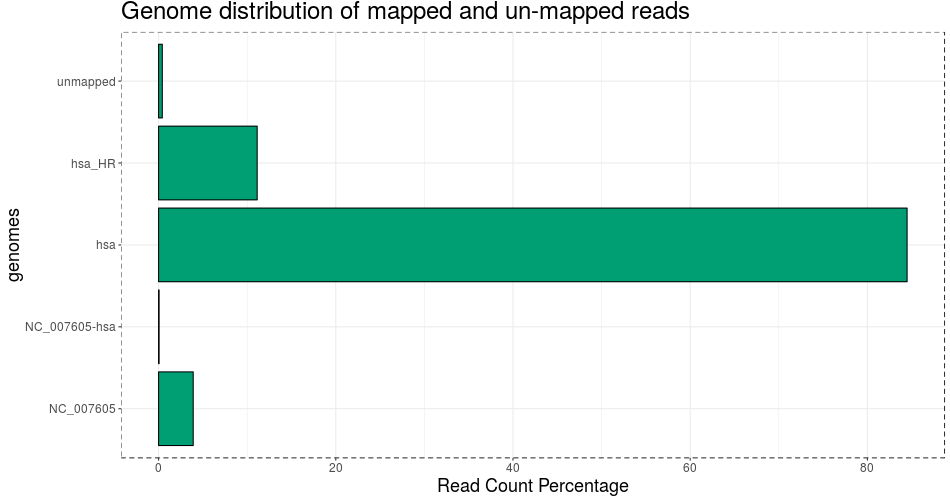
Used genome assembly bowtie1 index: GRCh38_p10_mp:NC_007605
---- ANNOTATIONS ----
Used miRNAs reference species from miRBase v22
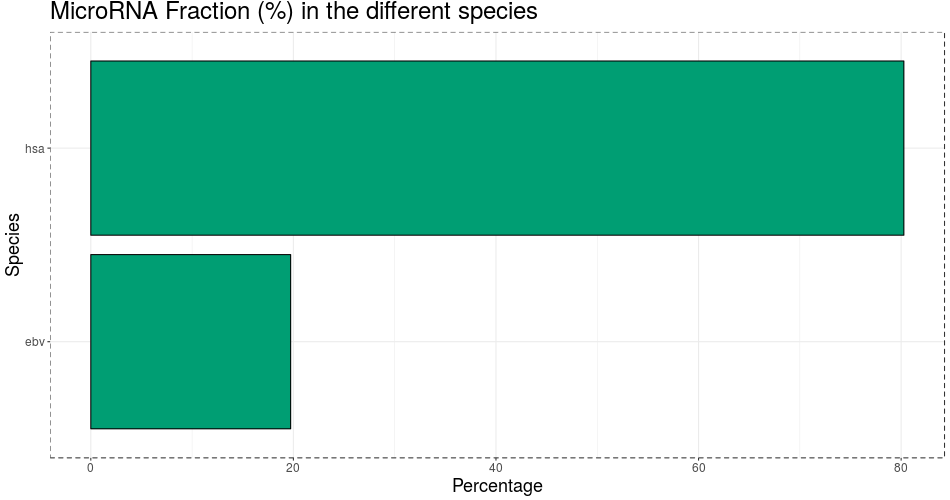
Used miRNAs for species: hsa:ebv
Used ncRNA annotations:
libs=GRCh38_p10_genomic_tRNA (Genomic tRNA database)
libs=hsa_yRNA (yRNAs)
libs=vRNA (vault RNAs)
libs=GRCh38_p10_ncRNA (Ensembl release 91 (ncRNA))
libs=GRCh38_p10_cdna (Ensembl release 91 (cDNA))
libs=GRCh38_p10_RNAcentral (RNAcentral database 7.0)
---- Preprocessing and adapter trimming parameters: ----
Unknown protocol: Illumina
---No quality filtering was used (only reads with ambigous bases are filtered out- ---
Number of Raw input reads : 6203008
Number of adapter trimmed reads: 6114811 (98.57815756484595)
--- Number of filtered reads ----
Quality Filtered Reads : 18872
Reads below min read count : 309386
Reads above max. length : 0
Reads below min. length : 1457621
Percentage of AdapterDimer : 3.4880611813859517
Reads in analysis: 4328932 (69.7876256164751)
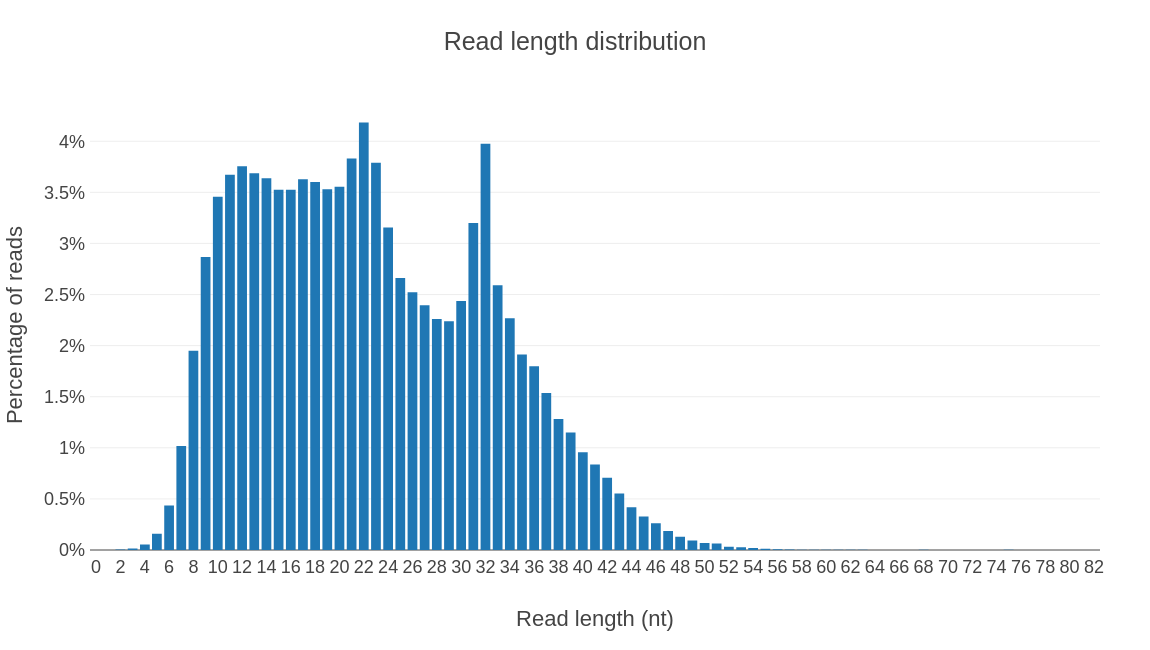
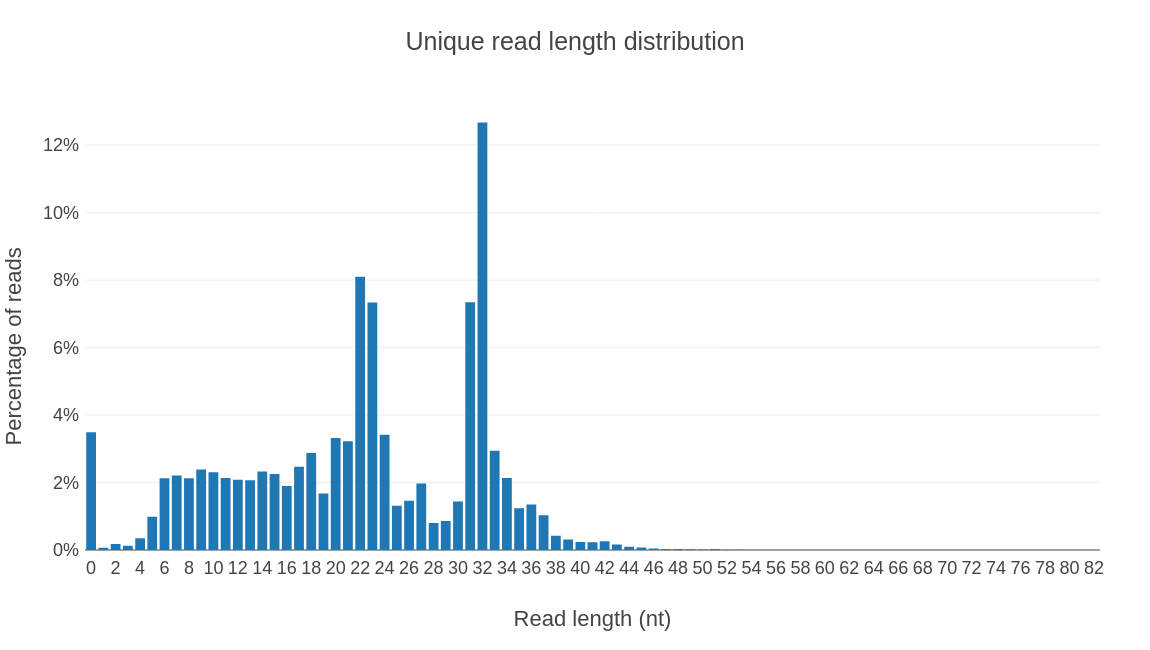
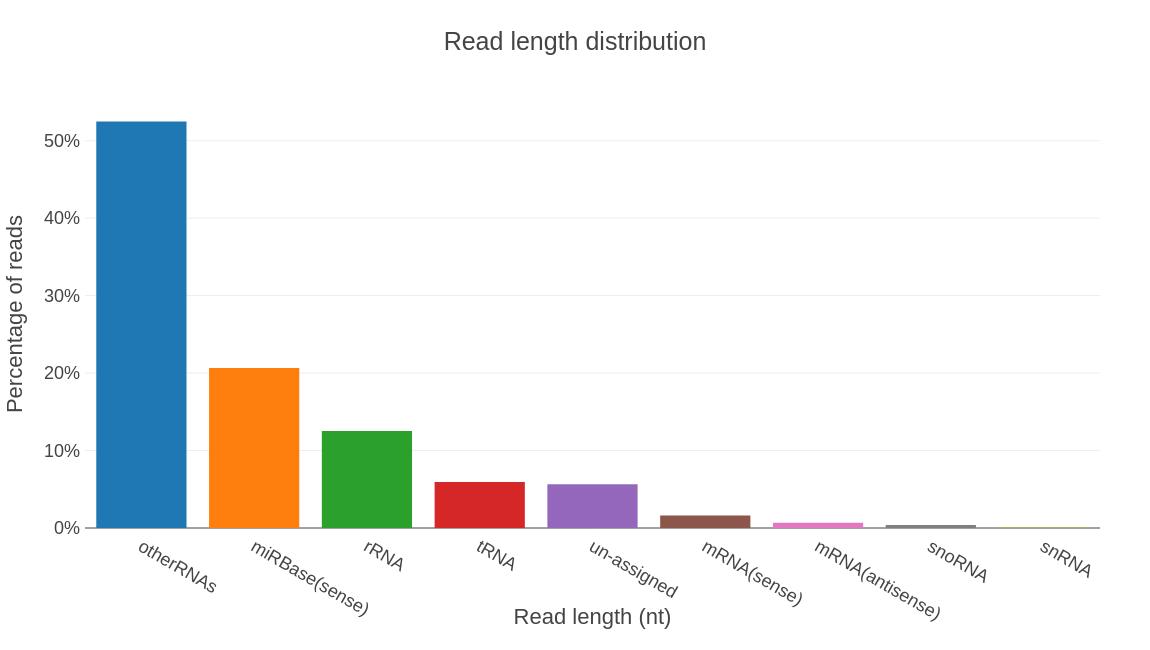
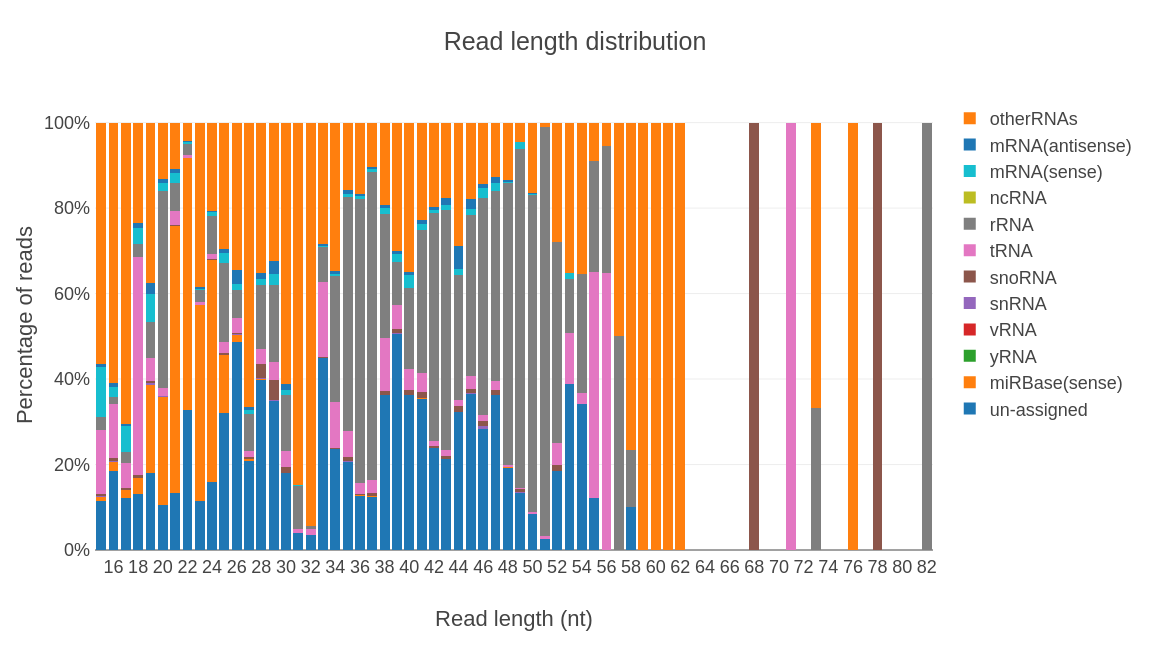
Profiling results by rna type.
| Library | Description | Orientation | UR | URperc | RC | RCperc | RPMassigned | RPManalysis |
|---|---|---|---|---|---|---|---|---|
| mature | miRBase v22 | antisense | 5 | 0.01 | 12 | 0 | 3.32 | 3.13 |
| hsa_yRNA | yRNAs | sense | 4391 | 5.38 | 1473394 | 38.48 | 407806.31 | 384815.32 |
| GRCh38_p10_genomic_tRNA | Genomic tRNA database | sense | 7711 | 9.44 | 225460 | 5.89 | 62402.87 | 58884.77 |
| GRCh38_p10_RNAcentral | RNAcentral database 7.0 | sense | 2839 | 3.48 | 466482 | 12.18 | 129112.99 | 121833.96 |
| GRCh38_p10_ncRNA | Ensembl release 91 (ncRNA) | antisense | 10576 | 12.95 | 143712 | 3.75 | 39776.64 | 37534.14 |
| GRCh38_p10_ncRNA | Ensembl release 91 (ncRNA) | sense | 18703 | 22.9 | 307527 | 8.03 | 85117.39 | 80318.71 |
| vRNA | vault RNAs | antisense | 2 | 0 | 6 | 0 | 1.66 | 1.57 |
| hairpin | -- | sense | 183 | 0.22 | 2390 | 0.06 | 661.5 | 624.21 |
| GRCh38_p10_genomic_tRNA | Genomic tRNA database | antisense | 82 | 0.1 | 599 | 0.02 | 165.79 | 156.44 |
| GRCh38_p10_cdna | Ensembl release 91 (cDNA) | sense | 6189 | 7.58 | 103094 | 2.69 | 28534.38 | 26925.69 |
| GRCh38_p10_cdna | Ensembl release 91 (cDNA) | antisense | 5882 | 7.2 | 89410 | 2.34 | 24746.92 | 23351.76 |
| hairpin | -- | antisense | 8 | 0.01 | 18 | 0 | 4.98 | 4.7 |
| mature | miRBase v22 | sense | 6525 | 7.99 | 788887 | 20.6 | 218348.31 | 206038.44 |
| hsa_yRNA | yRNAs | antisense | 1 | 0 | 2 | 0 | 0.55 | 0.52 |
| GRCh38_p10_RNAcentral | RNAcentral database 7.0 | antisense | 249 | 0.3 | 4318 | 0.11 | 1195.14 | 1127.76 |
| vRNA | vault RNAs | sense | 313 | 0.38 | 7664 | 0.2 | 2121.24 | 2001.65 |
| un-assigned | -- | --- | 18021 | 22.06 | 215859 | 5.64 | 0 | 56377.22 |
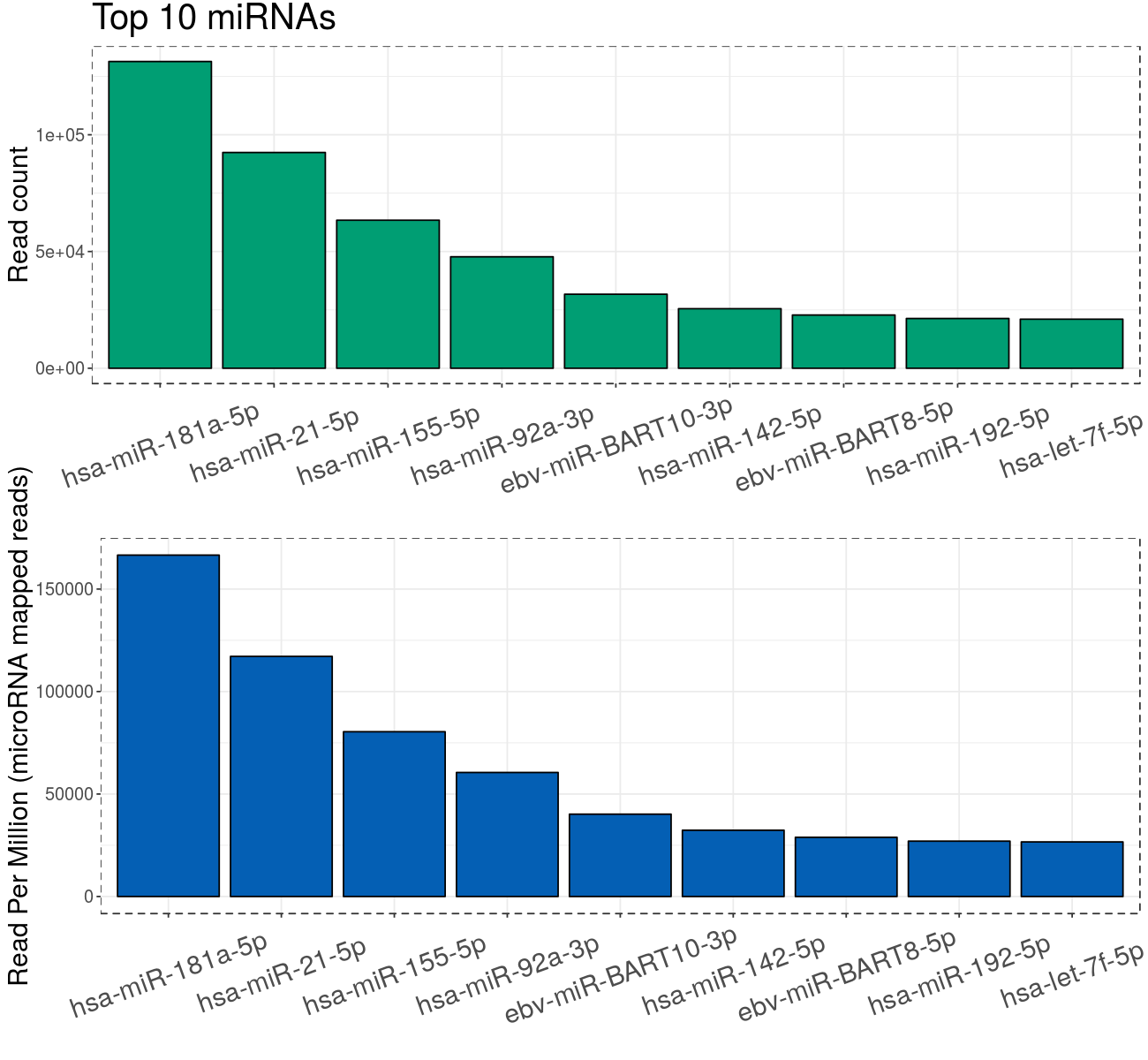
MiR profiling results
- Detected mature miR:
- 567 (21.0%)
- Reads mapped to miRBase hairpins:
- 2390 (0.07%)
- Reads mapped to miRNAs:
- 788887 (21.83%)
- Detected hairpin miR:
- 394 (20.29%)
- MicroRNA details:
- isomiR details:
vault RNAs
- Mapped reads in sense direction:
- 7664(0.21%)
- multiple assignment (MA)
- single assignment (SA)
- Mapped reads in antisense direction:
- 6(0.0%)
- MA antisense
- SA antisense
Genomic tRNA database
- Mapped reads in sense direction:
- 225460(6.24%)
- multiple assignment (MA)
- single assignment (SA)
- Mapped reads in antisense direction:
- 599(0.02%)
- MA antisense
- SA antisense
Ensembl release 91 (ncRNA)
- Mapped reads in sense direction:
- 307527(8.51%)
- multiple assignment (MA)
- single assignment (SA)
- Mapped reads in antisense direction:
- 143712(3.98%)
- MA antisense
- SA antisense
Ensembl release 91 (cDNA)
- Mapped reads in sense direction:
- 103094(2.85%)
- multiple assignment (MA)
- single assignment (SA)
- Mapped reads in antisense direction:
- 89410(2.47%)
- MA antisense
- SA antisense
yRNAs
- Mapped reads in sense direction:
- 1473394(40.78%)
- multiple assignment (MA)
- single assignment (SA)
- Mapped reads in antisense direction:
- 2(0.0%)
- MA antisense
- SA antisense
RNAcentral database 7.0
- Mapped reads in sense direction:
- 466482(12.91%)
- multiple assignment (MA)
- single assignment (SA)
- Mapped reads in antisense direction:
- 4318(0.12%)
- MA antisense
- SA antisense
- tRNA genes:
- details